We are broadly interested in how the molecular properties of viral proteins and antibodies constrain their evolution and co-evolution.
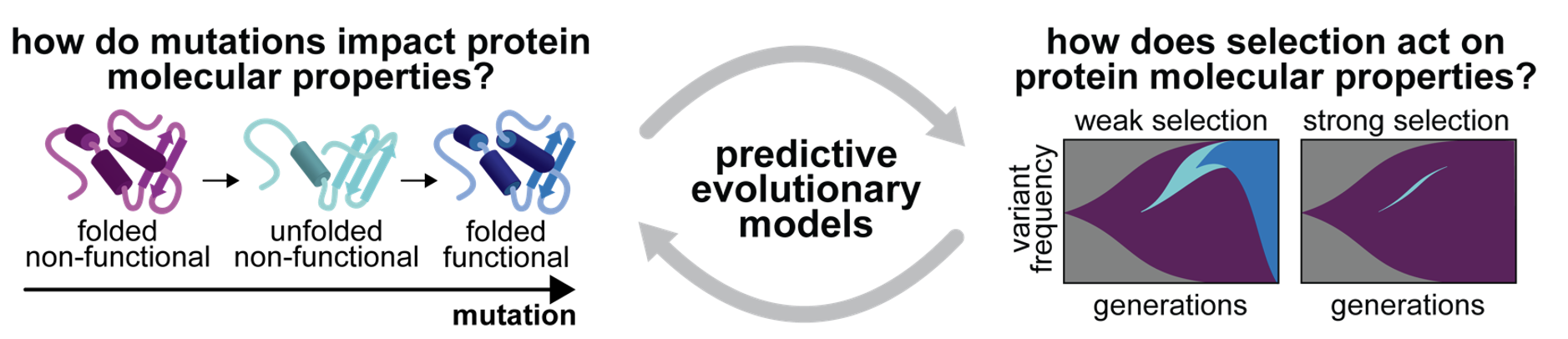
Viral proteins and antibodies acquire amino acid substitutions at a rate orders of magnitude above most eukaryotic proteins. These substitutions can have pleiotropic consequences on protein stability, folding, and function. Our lab is developing high-throughput evolution and phenotyping assays to determine how these properties, and trade-offs between them, constrain and potentiate the evolution of viral proteins and antibodies, and how this varies between distinct selection environments. These experimental platforms will enable us to (1) determine key constraints on protein evolution, (2) predict the emergence of new viral variants, and (3) design therapeutic strategies that are refractory to the development of resistance.
Ongoing projects in the lab include:
- Development of high-throughput methods for measuring biophysical properties of natively-synthesized proteins
Existing high-throughput protein phenotyping methods typically express the protein of interest in a system that lacks the native post-translational modification, folding, and processing machinery. This machinery critically impacts the stability, folding, and function of diverse proteins – especially viral proteins and antibodies – and motivates our work to develop new methods for proteins synthesized in human cells.
- Molecular mechanisms of viral evolution
The high mutation rate of RNA viruses like influenza, SARS-CoV-2, and HIV mediates evasion of the immune response and adaptation to new hosts. These mutations can also have a significant cost to viral protein stability and folding. Towards developing predictive models of viral evolution, we are examining the extent to which the stability, folding, and function of viral proteins shape the evolutionary dynamics of circulating viruses. We also employ experimental evolution and mutagenic libraries to dissect the contributions of specific mutations and selection pressures.
- Biophysical constraints of antibody affinity maturation
We’re interested in understanding how the adaptive immune system can mature antibodies that recognize diverse pathogens while avoiding non-productive interactions with self-proteins. To study this trade-off, we’re examining changes in the biophysical properties of the human antibody repertoire during somatic hypermutation. We’re especially interested in the biophysical basis of antibody evolvability and whether specific exposure regimens can shepherd the maturation of broadly protective antibodies.